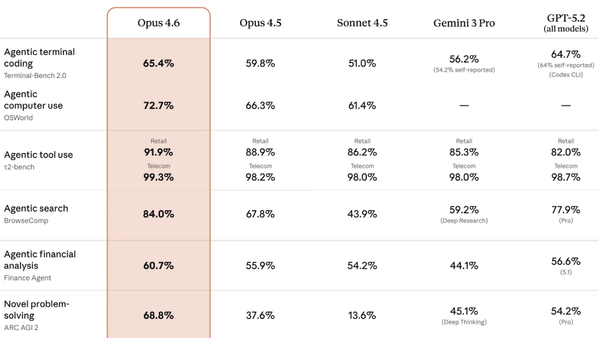
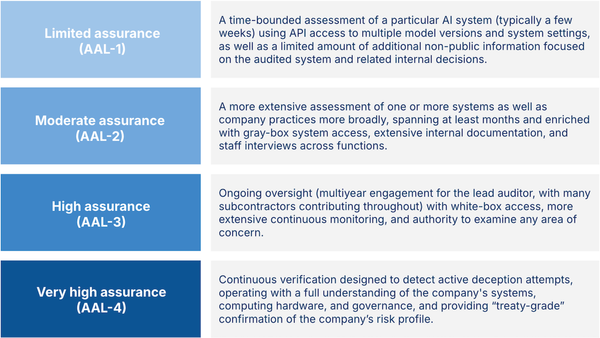
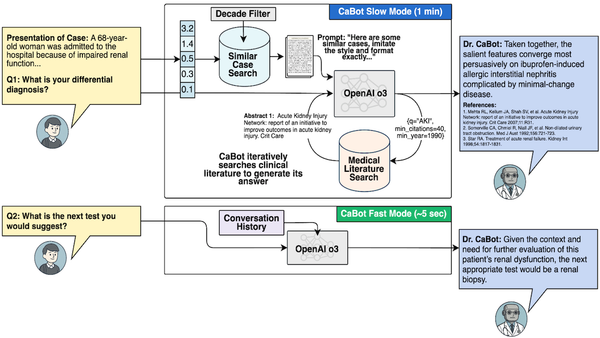
Multimodal Models for Biomedicine by Pengtao Xie: Pengtao Xie of UC-San Diego on why medical models need to visualize tiny chemicals and large organs
Over the past few years, we have seen rapid progress in models that jointly reason over text, images, sequences, graphs, and time series. Yet in biomedical settings, these capabilities often remain fragmented, brittle, or difficult to interpret.

Over the past few years, we have seen rapid progress in models that jointly reason over text, images, sequences, graphs, and time series. Yet in biomedical settings, these capabilities often remain fragmented, brittle, or difficult to interpret. In 2026, I hope the community moves decisively toward building multimodal models that are not only powerful but also scientifically grounded, transparent, and genuinely useful to biomedical discovery and clinical decision-making.
A key priority should be deep multimodal integration, rather than superficial concatenation of modalities. Biological systems are inherently multi-scale and multi-view: molecules, cells, tissues, organs, and patients are connected through complex mechanisms that span sequences, structures, images, and longitudinal records. Foundation models should reflect this structure by learning aligned representations that preserve biological meaning across modalities, enabling coherent reasoning from molecular mechanisms to phenotypic outcomes. Achieving this will require new pretraining objectives, better inductive biases, and principled ways to encode biological context. For instance, researchers could design objectives that explicitly align representations across modalities using shared biological anchors such as pathways, cell states, or disease phenotypes, so information learned from one view remains meaningful when transferred to another.
Another critical focus is interpretability. In biomedicine, predictions alone are rarely sufficient. Researchers and clinicians need to understand why a model makes a decision, what evidence it relies on, and how its outputs relate to known biology. As multimodal models grow larger and more general, the AI community should prioritize explanation methods that operate across modalities, allowing users to trace predictions back to molecular interactions, image regions, or temporal patterns in patient data. For instance, models could be designed to produce explanations as structured cross-modal attributions, explicitly linking elements in one modality (e.g., genes or residues) to evidence in another (e.g., image regions or time points).
Data efficiency and adaptability should be central goals. Many biomedical domains suffer from limited labeled data, strong distribution shifts, and incomplete knowledge. Multi-modal foundation models must be able to adapt to new tasks, diseases, and institutions with minimal retraining, while maintaining robustness and calibration. Parameter-efficient adaptation, continual learning, and uncertainty-aware inference are especially important in this context.
Finally, progress in 2026 should be measured not only by benchmarks, but also by integration into biomedical workflows. This includes tools that support hypothesis generation, experimental design, and interactive exploration. Such tools would allow domain experts to engage in multi-turn dialogue with models rather than passively consume predictions.
If the AI community aligns around these priorities, multimodal foundation models could become trusted partners in biomedical research, accelerating understanding while respecting the complexity and responsibility inherent in supporting human health.
Pengtao Xie is an associate professor at UC San Diego and an adjunct faculty member at Mohamed bin Zayed University of Artificial Intelligence in Abu Dhabi. Previously, he was senior director of engineering at Petuum, a generative AI startup.